π-HuB Project Core Platform Establishes High-Efficiency Cell-Type-Resolved Spatial Proteomics Technology
03
2025
The localization and interactions of proteins are fundamental to their biological functions, making it critical to understand their roles within cells and tissues. Spatial proteomics technology enables high-resolution, in-depth protein analysis while preserving the spatial integrity of cells and tissues, thereby advancing our understanding of intracellular signaling, intercellular communication, and interactions with the extracellular matrix. Recognized as the Nature Methods 2024 Method of the Year, spatial proteomics has demonstrated its revolutionary potential in unraveling biological complexity and advancing precision medicine and cancer research.

In March 2025, a collaborative team from the Guangdong International Academy of Intelligent Medicine, the He Fuchu Team of the Wise-eye Large-Scale Facility Project, the Chao Liu Research Group at Beihang University, and Dr. Honghua Zhang from Sun Yat-Sen Memorial Hospital published a study titled "High-Efficiency Cell-Type Proteomics Strategy Deciphers Cholangiocarcinoma Fibrosis-Associated Pathological Heterogeneity" in Analytical Chemistry. The study describes a high-efficiency cell-type-resolved spatial proteomics technology that integrates the pathological image analysis algorithm π-CellSeg-CCA, laser microdissection (LMD), the SISPROT sample preparation technique, and high-sensitivity mass spectrometry. This approach was successfully applied to dissect the pathological heterogeneity of cholangiocarcinoma (CCA) fibrosis.
The π-CellSeg-CCA algorithm automatically annotates CCA and normal bile duct cell clusters in pathological images with 90% accuracy, then exports their contours for efficient LMD-based dissection. Using this method, the team identified over 8,000 proteins from just 1 mm² of tissue, including CCA-specific biomarkers. Notably, they discovered MUC16, a protein specifically upregulated in fibrotic CCA regions, and revealed its association with poor patient prognosis and disease progression.
Key Findings
01. Development of High-Efficiency Cell-Type-Resolved Spatial Proteomics Technology
The study established a spatial proteomics workflow combining π-CellSeg-CCA, LMD, SISPROT, and timsTOF Pro mass spectrometry (Figure 1). After H&E staining and whole-slide imaging, π-CellSeg-CCA automatically identifies and segments CCA/normal bile duct cell clusters, guiding LMD-based dissection. The collected samples are processed via SISPROT and analyzed by high-sensitivity mass spectrometry.
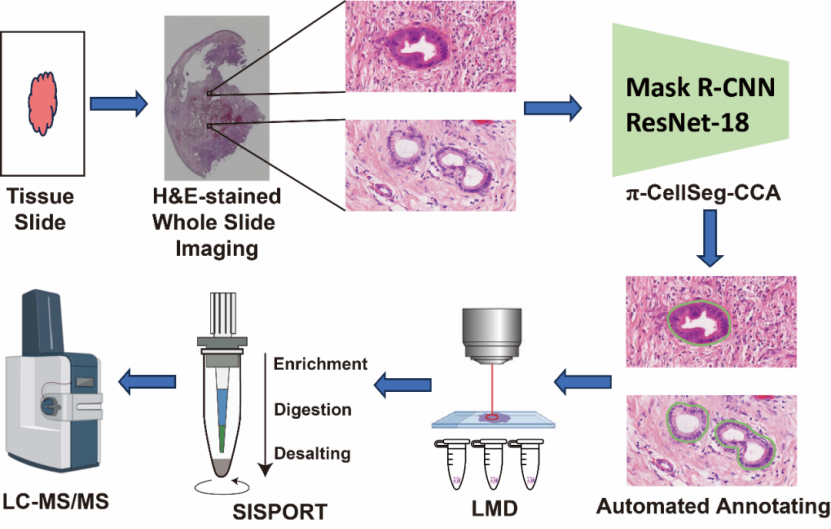
Figure 1. High-efficiency cell-type-resolved spatial proteomics technology.
02. Development and Validation of π-CellSeg-CCA Algorithm
The π-CellSeg-CCA algorithm leverages Mask R-CNN-DCT (enhanced with a discrete cosine transform module for high-resolution mask generation) and ResNet-18 for classification, achieving 90% accuracy in distinguishing malignant from normal bile duct clusters (Figure 2A). Figure 2B showcases examples of high-confidence CCA (99% probability) and normal bile duct classifications.
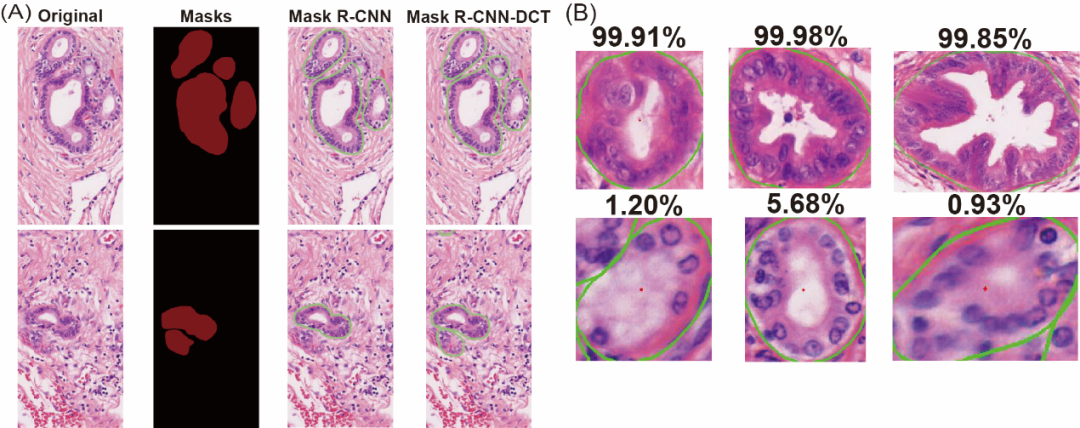
Figure 2. Performance of π-CellSeg-CCA in (A) segmentation and (B) classification (percentage indicates malignancy probability).
03. Cell-Type Proteomic Analysis of CCA Heterogeneity
The team analyzed 6 normal bile duct (Normal BD), 5 non-fibrotic CCA (NF CCA), and 7 fibrotic CCA (Fibro CCA) samples (Figure 3A), yielding the following insights:
u 8,000+ proteins were identified per 1 mm² sample, with high reproducibility (Pearson’s r > 0.85) (Figure 3B, C).
u Known CCA biomarkers (S100P, MUC1, MUC5AC) were confirmed, validating the method’s accuracy (Figure 3D).
u MUC16 and S100A2 were uniquely upregulated in fibrotic CCA (Figure 3E).
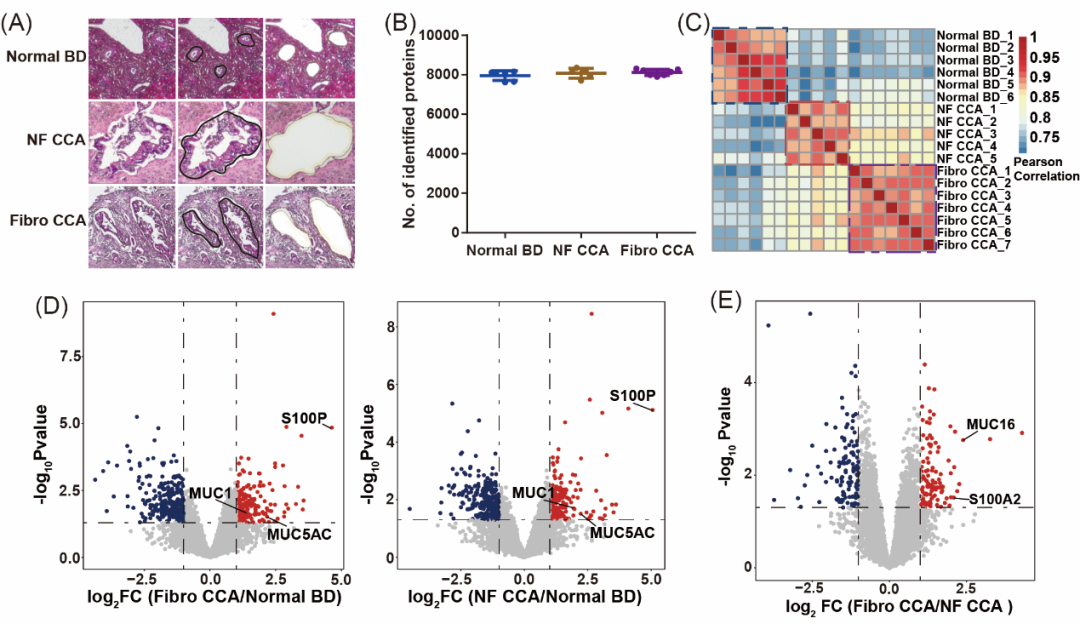
Figure 3. Cell-type proteomic analysis of CCA heterogeneity.
04. Validation of MUC16’s Role in Fibrotic CCA Progression
Follow-up experiments demonstrated:
u MUC16 is specifically overexpressed in fibrotic CCA and correlates with fibrosis severity (Figure 4A, B).
u Clinically, high MUC16 expression is linked to advanced T-stage, lymph node metastasis (LNM), and distant metastasis (DM) (Figure 4C, D).
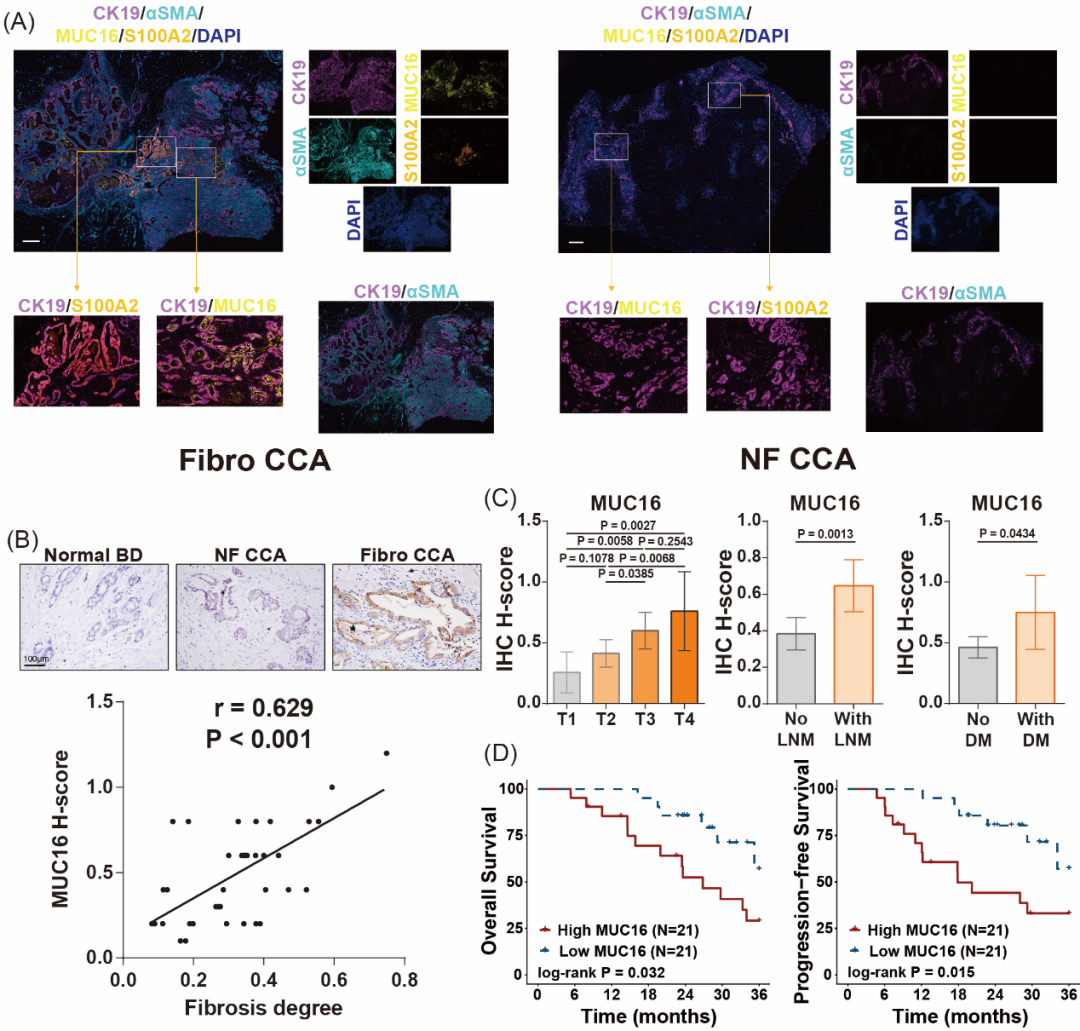 Figure 4. Validation of MUC16’s association with fibrotic CCA progression.
Figure 4. Validation of MUC16’s association with fibrotic CCA progression.
Conclusion
This study developed a high-efficiency spatial proteomics platform that deciphers CCA fibrosis heterogeneity. The π-CellSeg-CCA algorithm significantly enhances analytical efficiency and is adaptable to other cancers, offering broad applications in precision medicine.
Corresponding Authors:
Fuchu He (Guangdong International Academy of Intelligent Medicine; Wise-eye Large-Scale Facility Project)
Wendong Chen (Guangdong International Academy of Intelligent Medicine)
Chao Liu (Beihang University)
Co-First Authors:
Zhiyang Su (Southern University of Science and Technology-Wise-eye Joint Program)
Honghua Zhang (Sun Yat-Sen Memorial Hospital)
Funding: Supported by the π-HuBproject and the Wise-eye Large-Scale Facility Pre-Research Project.
Original Article: https://doi.org/10.1021/acs.analchem.4c06106
Wise-eye Spatial Proteomics Service Platform Announcement
The Wise-eye Spatial Proteomics Service Platform now offers global research institutions, hospitals, and enterprises end-to-end spatial proteomics services, including:
Cell-type spatial proteomics
Holographic spatial proteomics
Spatial phosphoproteomics
Platform Equipment:
Automated tissue processors, microtomes, cryostats
Leica LMD7 laser microdissection system
High-throughput SISPROT sample preparation
High-sensitivity mass spectrometry
Contact:
Tel: 020-66312101 ext. 8303
Email: 13522169171@163.com (Dr. Li), chenwdscib@163.com (Dr. Chen)
Address: Building A, 83 Ruihe Road, Huangpu District, Guangzhou


