Cell: π-HuB Project member Collaborates to Establish Single-Cell Protein Turnover Rate Analysis Technology
04
2025
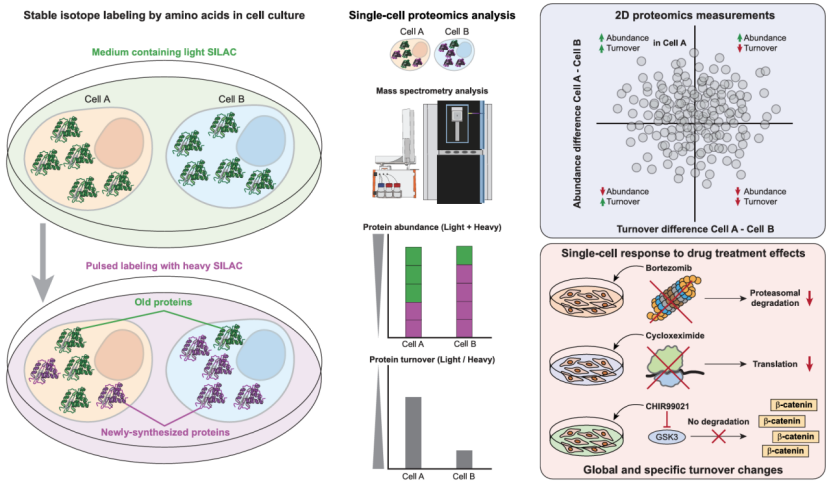
On March 31, 2025, a collaborative study led by Professor Jesper Olsen’s team at the University of Copenhagen and Dr. Zilu Ye’s team at the Chinese Academy of Medical Sciences/Suzhou Institute of Systems Medicine was published online in Cell under the title "Global Analysis of Protein Turnover Dynamics in Single Cells." The research developed an advanced single-cell proteomics technology called SC-pSILAC (Single-Cell Pulsed Stable Isotope Labeling by Amino Acids in Cell Culture), enabling for the first time the simultaneous precise measurement of protein abundance and turnover rates at the single-cell level. This breakthrough provides a novel dimension and perspective for studying cellular heterogeneity.

Protein Turnover: A Key to Cellular Function and Heterogeneity
Protein synthesis and degradation (i.e., protein turnover) are critical processes for maintaining cellular function and responding to environmental changes. However, traditional proteomics analyses are typically conducted at the bulk cell level, which masks significant heterogeneity and dynamic variations among individual cells due to population averaging. Although single-cell proteomics has advanced significantly in recent years, sensitivity limitations have hindered precise resolution of protein turnover rates in single cells.
To overcome this bottleneck, the researchers integrated pulsed stable isotope labeling (pSILAC) with their previously developed high-throughput, high-sensitivity Chip-Tip single-cell proteomics technology
(see Nature Methods: https://www.nature.com/articles/s41592-024-02558-2).
This innovation allowed them to achieve single-cell protein turnover studies for the first time. The team successfully quantified over 4,000 proteins in 10-cell pools and 3,000 proteins in individual HeLa cells, with the majority of proteins simultaneously labeled by light and heavy isotopes. This milestone marks the first precise mapping of protein synthesis and degradation dynamics at the single-cell level.
SC-pSILAC: Technology and Applications
The SC-pSILAC technology combines:
pSILAC labeling: Cells are switched from light (non-isotopic) to heavy (isotopic) amino acid media, enabling tracking of protein synthesis and degradation.
High-sensitivity mass spectrometry: Leveraging the Orbitrap Astral platform and narrow-window DIA (nDIA) for multiplexed quantification.
Single-cell resolution: Detects both light and heavy labels in individual cells, revealing turnover heterogeneity.
Key Findings:
Drug Response Profiling: SC-pSILAC sensitively captured drug-induced changes in protein turnover. For example, GSK3 inhibitor CHIR99021 treatment increased β-catenin (CTNB1) abundance while slowing its degradation, validating the Wnt pathway’s regulatory mechanism.
Stem Cell Differentiation: Over 1,000 single cells were analyzed across six stages of induced pluripotent stem cell (iPSC) differentiation spanning two months. Protein turnover rates revealed functional distinctions invisible to traditional abundance measurements, such as:
Core histones: Turnover rates distinguished dividing from non-dividing cells, with faster turnover in proliferating iPSCs.
Cell Size Scaling: Histones and cell-cycle proteins did not scale with cell size, while cytoskeletal proteins exhibited positive correlations.
Protein Complex Coordination: Members of complexes like the proteasome and ribosome showed co-regulated turnover, highlighting functional modularity.
Implications and Future Directions
This study transcends the limitations of static single-cell proteomics by introducing dynamic turnover measurements, offering unprecedented tools to investigate:
Cell differentiation mechanisms
Post-transcriptional regulation
Disease-associated protein dysregulation (e.g., cancer dormancy, neurodegenerative disorders)
The technology’s ability to resolve single-cell heterogeneity positions it as a transformative resource for precision medicine and drug discovery, particularly in identifying therapeutic targets modulating protein turnover.
Authors and Funding
First & Co-Corresponding Author: Dr. Pierre Sabatier (Uppsala University/University of Copenhagen)
Corresponding Authors: Dr. Zilu Ye (Chinese Academy of Medical Sciences) and Prof. Jesper Olsen (University of Copenhagen)
Support: π-HuB Project and Novo Nordisk Foundation
Original Article: https://doi.org/10.1016/j.cell.2025.03.002

